Pages:
Example CompariMotif Analysis
A typical application for CompariMotif is given in the SLiMFinder paper (Edwards, Davey & Shields 2007, Example 1), in which HPRD interaction datasets for 14-3-3 proteins were analysed using SLiMFinder, returning several significant motifs (p<0.05, see below). NB. The Input Help Page has these motifs in the recommended CompariMotif input format, although raw SLiMFinder and SLiMDisc delimited text results can be loaded directly into CompariMotif for analysis without any reformatting.
SLiMFinder results for 14-3-3 interaction datasets from HRPD. reproduced from Edwards, Davey & Shields 2007 (Table 2).
| Beta/Alpha (YWHAB) | 220 (55) | - | 0.0% |
| Epsilon (YWHAE) | 117 (34) | R..S.P..L * | 40.0% |
| Eta (YWHAH) | 83 (27) | GR.[ST]..P * | 37.0% |
| Gamma (YHWAG) | 383 (101) | ^.[AS][AGS] *** | 40.6% |
| KE..K * | 35.6% | ||
| Sigma (SFN) | 48 (21) | - | 0.0% |
| Theta/Tau (YWHAQ) | 132 (42) | P..P..P * | 66.7% |
| Zeta/Delta (YWHAZ) | 190 (58) | [AGS]..P..P..P *** | 48.3% |
| ^.[AGS][GS] ** | 27.6% | ||
| FR..[ST].S ** | 19.0% | ||
| [ST]P.[ST]P * | 34.5% | ||
| Y.C.PG.L * | 6.9% |
bNumber of proteins in dataset. Number of UPC is given in brackets.
cThe most significant motif of each "cloud" returned by SLiMFinder. *p<0.05, **p<0.01, ***p<0.001.
dThe percentage of the dataset's UPC covered by occurrences of returned motifs in the same motif cloud.
These motifs were compared to the January 2008 version of the ELM database (Puntervoll et al. 2003) using CompariMotif with a "normalised IC" cut-off of 0.4 (Figures 1&2, below) Results were constrained such that fixed positions in an ELM must match a fixed position in the SLiMFinder motif.
In total, eight out of ten SLiMFinder motifs had matches with seventeen ELMs. The eight motifs with matches fell into three main clusters: (1) Three motifs matching known 14-3-3 motifs, (2) Three motifs matching SH3 binding motifs, and (3) Two motifs matching the highly degenerate LIG_PCNA_1 motif. In addition to the 14-3-3 and SH3 ELMs, matches to five phosphorylation ELMs were also identified; phosphorylation of the 14-3-3 motif is important for ligand recognition.
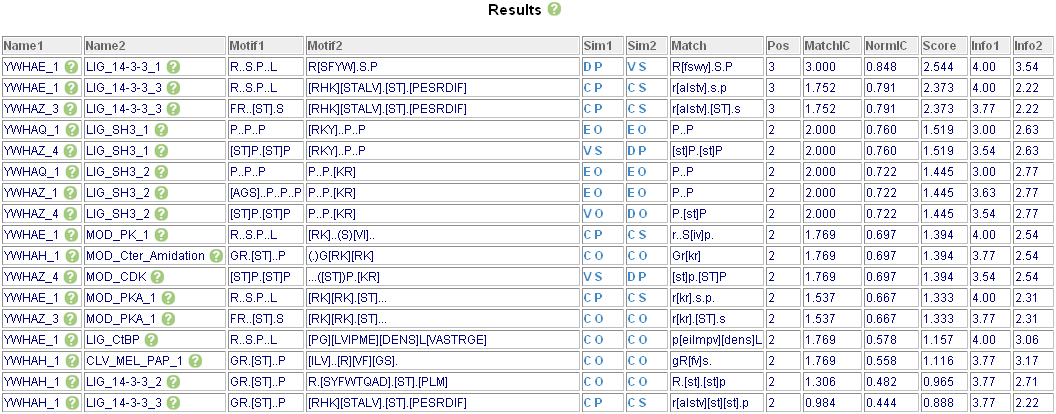
Figure 1. CompariMotif webserver main table output.

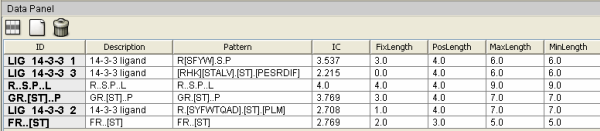
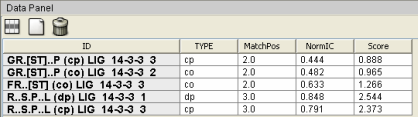
Figure 2. CompariMotif XGMML output visualized with Cytoscape (Shannon et al. 2003) (recoloured). Motifs returned by SLiMFinder analysis of 14-3-3 interaction datasets are shown as blue ellipses. ELMs with CompariMotif matches are shown as rectangles. These are pale red by default but the following groups have been manually recoloured: 14-3-3 ligands, green; SH3 ligands, orange, phosphorylation motifs, yellow. Arrows proceed from parent to subsequence motifs (bidirectional where equal); Data Panel details for 14-3-3 ELMs and connected nodes and edges are shown.
References
- Edwards RJ, Davey NE & Shields DC (2007). "Slimfinder: A probabilistic method for identifying over-represented, convergently evolved, short linear motifs in proteins." PLoS ONE 2(10): e967.
- Mishra GR et al. (2006). "Human protein reference database--2006 update." Nucleic Acids Res. 34(Database issue): D411-4.
- Puntervoll P et al. (2003). "Elm server: A new resource for investigating short functional sites in modular eukaryotic proteins." Nucleic Acids Res. 31(13): 3625-30.
- Shannon P et al. (2003). "Cytoscape: A software environment for integrated models of biomolecular interaction networks." Genome Res 13(11): 2498-504.