Pages:
How CompariMotif Works
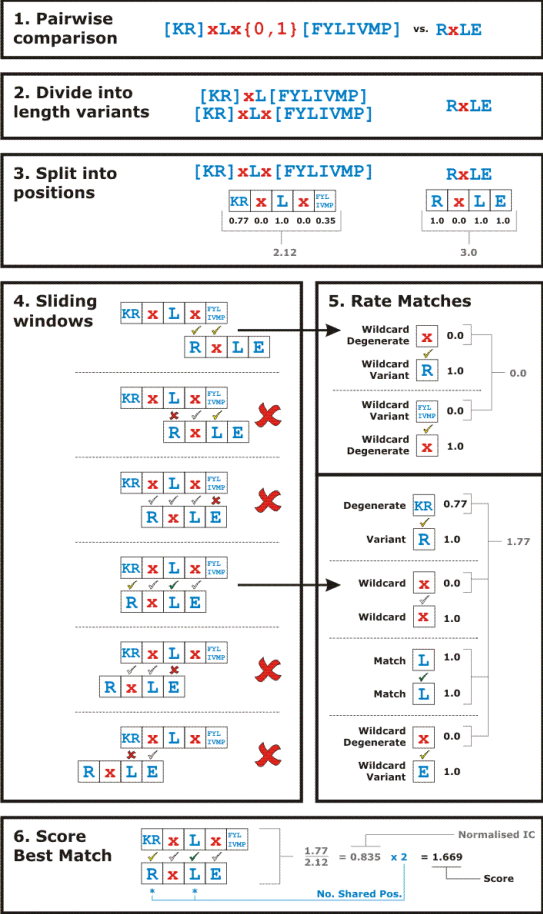
Overview of CompariMotif. See text below for details.
An overview of CompariMotif is shown above. Motifs are first compared for precise matches. If these are not found, then CompariMotif adopts a sliding window comparison in which every possible overlap between (variants of) the two motifs are compared against each other. Matches must meet a minimum match requirement determined by the minshare=X, normcut=X and matchfix=X options (see Options). Fixed positions in motifs are often more important that ambiguous ones, especially when the motif has been experimentally determined. For this reason, it is also possible to stipulate that all fixed positions in one or other motif (or both) match exactly to fixed positions in the compared motifs. This is controlled using the matchfix=X option.
Single Position Comparisons
For every comparison, each position in each motif is then rated according to its relationship with the compared position in the other motif:
- Match = perfect fixed position match
- Wildcard = wildcard in both motifs
- Wildcard variant = wildcard in compared motif but not in focal motif
- Wildcard degenerate = wildcard in focal motif but not in compared motif
- Ambiguous Match = ambiguities in both motifs comparing the same amino acids
- Degenerate Ambiguity = ambiguity in both motifs but the compared site is a subset of the ambiguity in the focal site
- Variant Ambiguity = ambiguity in both motifs but the focal site is a subset of the ambiguity in the compared site
- Degenerate = ambiguity in focal motif but fixed position in compared motif
- Variant = a fixed variant in the focal motif of a degenerate position in the compared motif
- Overlapping ambiguity = ambiguity in both motifs where 1+ amino acids overlap but each ambiguity also contains amino acids not in the other
- Bad ambiguity = ambiguity in focal motifs sharing no amino acids in common with compared motif
- Ambiguity mismatch = fixed position in focal motif that does not match ambiguity in compared motif
- Mismatch = different fixed positions in each motifs
Each positional comparison is then given an information content (IC) rating, if it is a "good" match. This is the lower IC out of the two positions compared. E.g. a fixed variant matching an ambiguity will take the IC of the ambiguity.
Selecting Pairwise Matches
The entire pairwise comparison is then rated for:
- Number of matching positions, allowing for degeneracy
- Number of exactly matching fixed positions
- Match Information content (IC), which is the sum of IC over all matched positions
- Number of incompatible positions (e.g. Bad ambiguities and mismatches)
The comparison is then rejected as a potential match if one of the following conditions is met:
- There are any incompatible positions. (If the mismatches=X option is used, this is relaxed.)
- The number of matched positions is less than that stipulated by minshare=X.
- The matchfix=X option is used and the relevant motif(s) in the comparison does not have exact matches at all its fixed positions.
- The normalised IC is below that set by normcut=X.
When a motif has multiple length variants and/or "NofM" elements (see Manual), each possible variant is compared.
Multiple variants and/or sliding windows can produce multiple matches that meet the acceptance criteria. In this case, the match with the best information content is used. In the case of tied information content, matches are assessed by the number of matching positions and then the number of exactly matching fixed positions. The earlier comparison made is considered "best" if all these stats tie.
Defining Motif Relationships
The best match is then considered to define the CompariMotif Relationships between the two motifs.