Pages:
Screenshot Walkthrough:
Server Frontpage
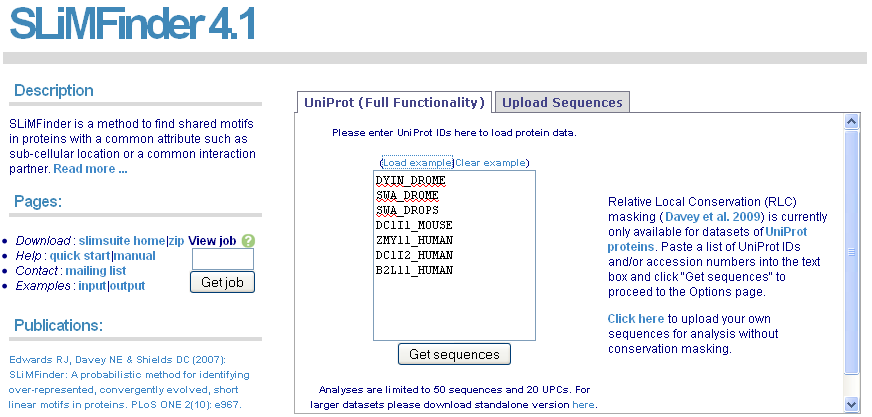
For full functionality, including conservation masking, input a list of UniProt IDs or accession numbers and click "Get Sequences". If you have already run SLiMFinder on a dataset, you can input the job ID and click "Get job" to jump straight to the results.
Input Dataset
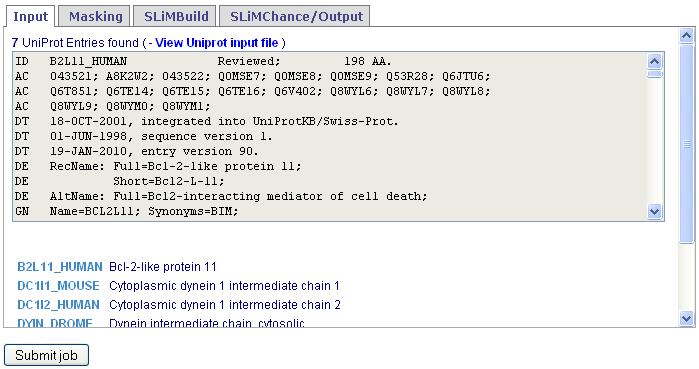
Downloaded UniProt entries will be visible in the text box and available to save for later viewing/re-use.
Alternative input entry
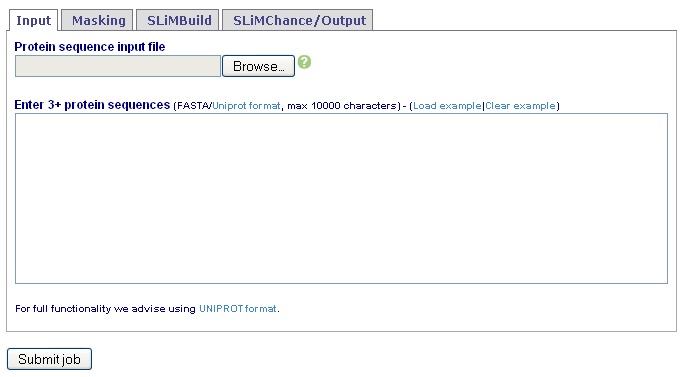
Alternatively, sequences can be directly pasted into the text box or a file uploaded. Fasta or UniProt formats are accepted.
Input Dataset
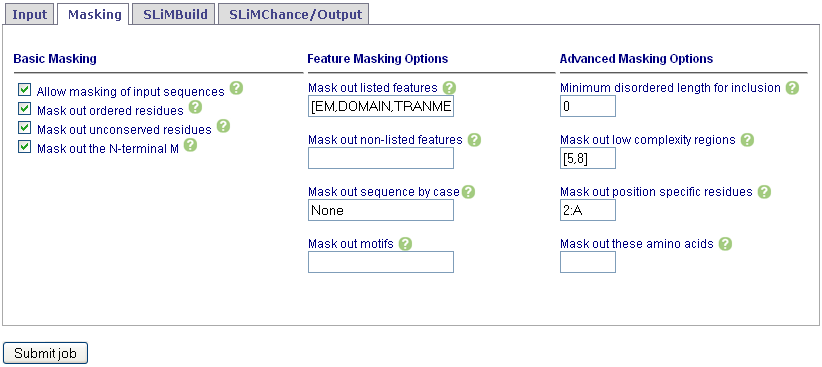
Set masking options. See Help for details. Note that consmask is only available for sequences downloaded directly from UniProt.
Input Dataset
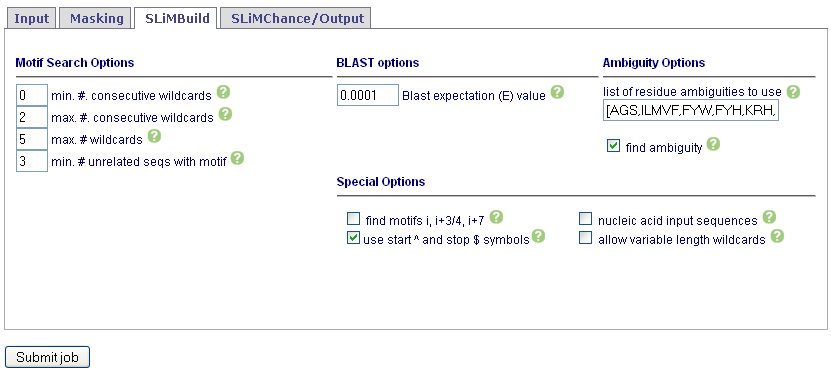
Set SLiMBuild options. See Help for details. To return motifs with degenerate wildcards, make sure wildvar is checked.
Input Dataset
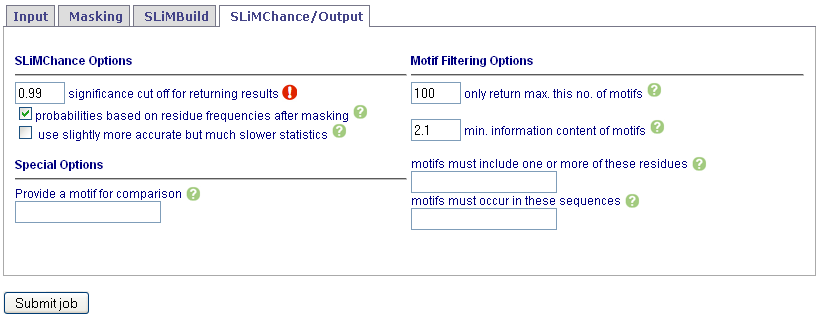
Set SLiMChance and Output options. Note that by default, the top 100 motifs with Sig < 1.0 are returned.
Results are unlikely to be genuine motifs unless Sig < 0.05. See Help for details.
To use the more accurate but slower References and CitationsSigV statistic make sure sigv is checked.
Once all options are set, click "Submit job".
Input Dataset
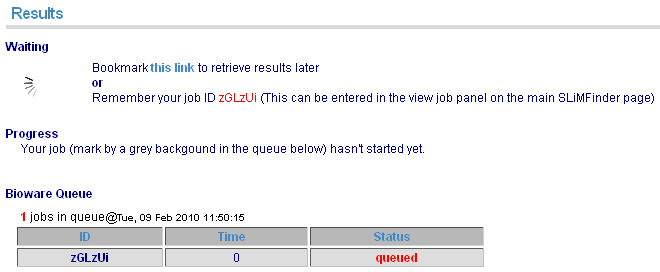
When your job is submitted, it will enter the bioware queue. The page will update once it starts running. Alternatively, the page can be bookmarked to revisit later.
Running job
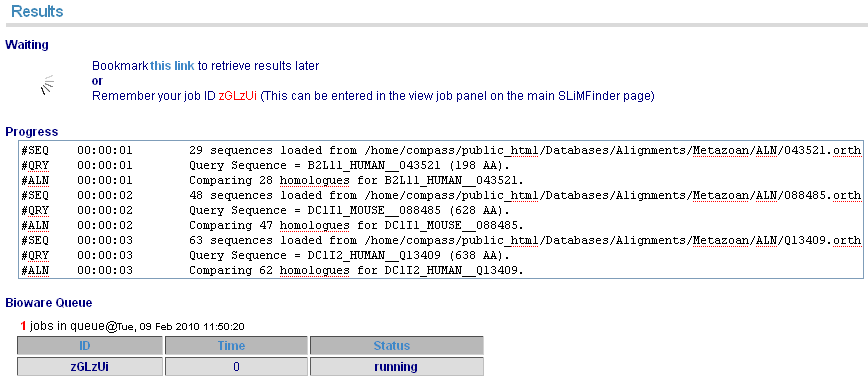
When your job is running, details of the log and current queue can be monitored. Alternatively, the page can be bookmarked to revisit later.
Results page
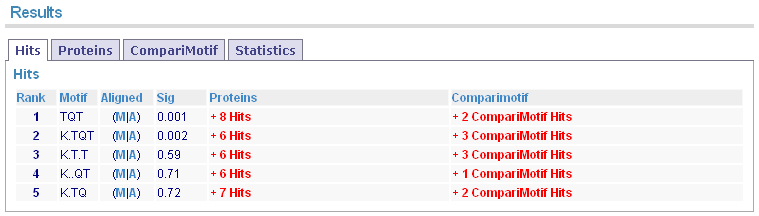
The initial results page shows summary results for returned motifs. Remember to check the "Sig" column as this indicates the estimated significance level of the motifs.
Expanded Hits
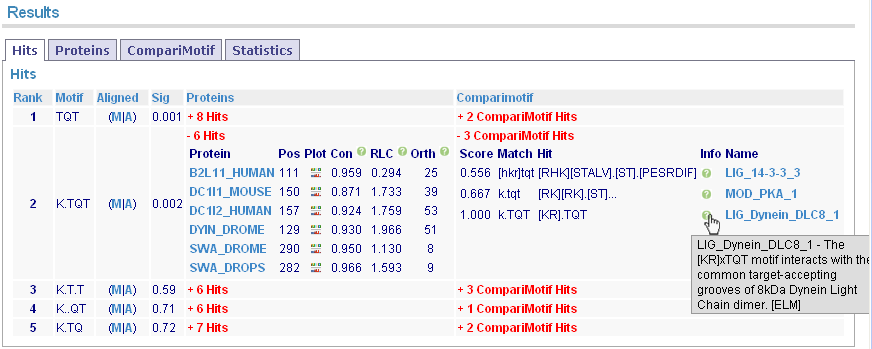
Clicking the red links expands details for the proteins and CompariMotif hits.
Motif alignments
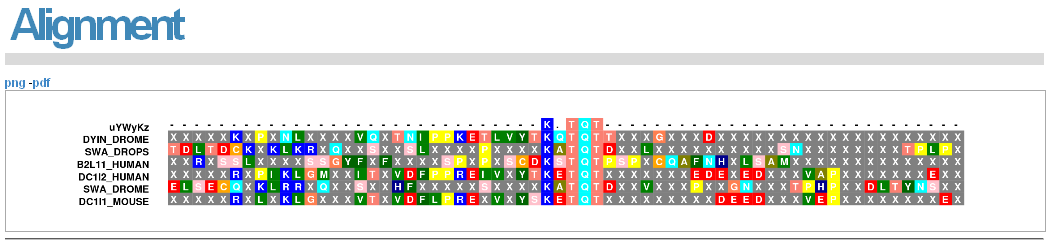

The "M" and "A" alignment links will visualise the motif in the input proteins, masked and unmasked respectively. These alignments can be saved by clicking on "png" or "pdf".
Protein orthologue alignments
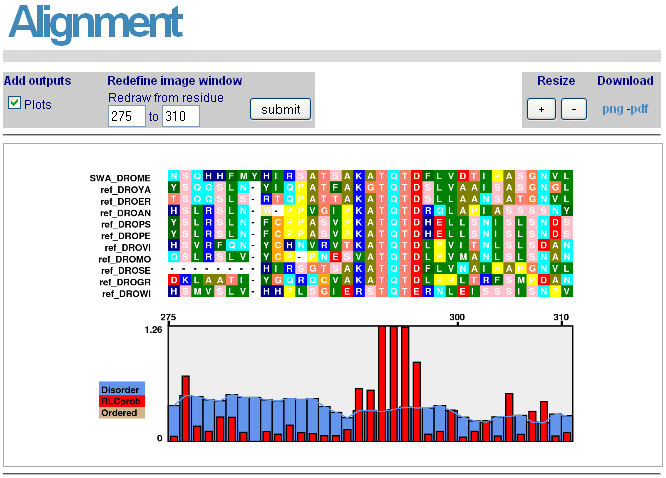
Alignments for each protein and its GOPHER orthologues around the motif of interest can be accessed by clicking on the "Plot" link for each protein/motif pair. Protein disorder and RLC scores are also visualised. The region can be altered to zoom in or out as desired. These alignments can be saved by clicking on "png" or "pdf".
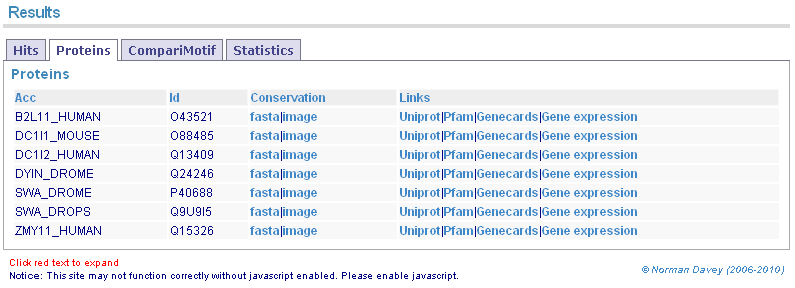
GOPHER orthologue alignments and links for input proteins.
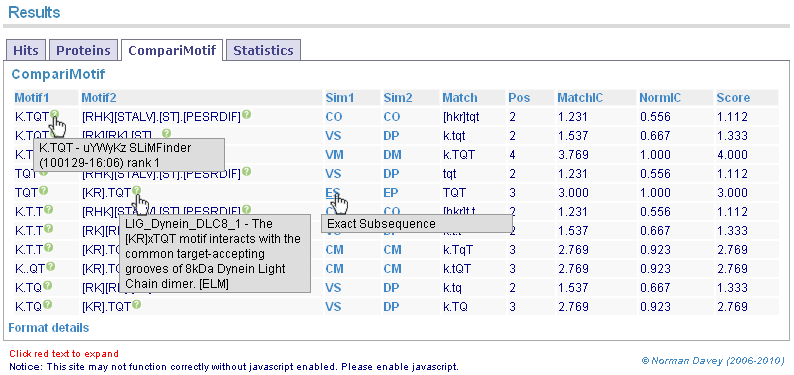
Returned motifs are compared to known literature motifs using CompariMotif. Mousing over key details gives more information.
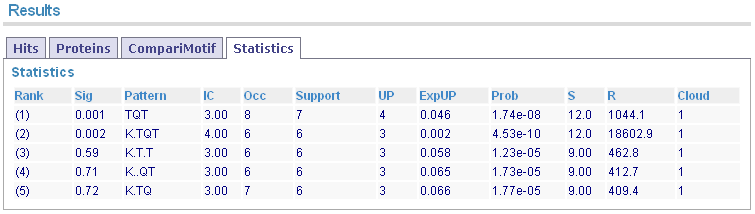
Summary statistics for each motif are displayed in a sortable table.

Raw data and additional visualisations may be downloaded from the panel on the left.