Pages:
Screenshot Walkthrough:
Server Frontpage
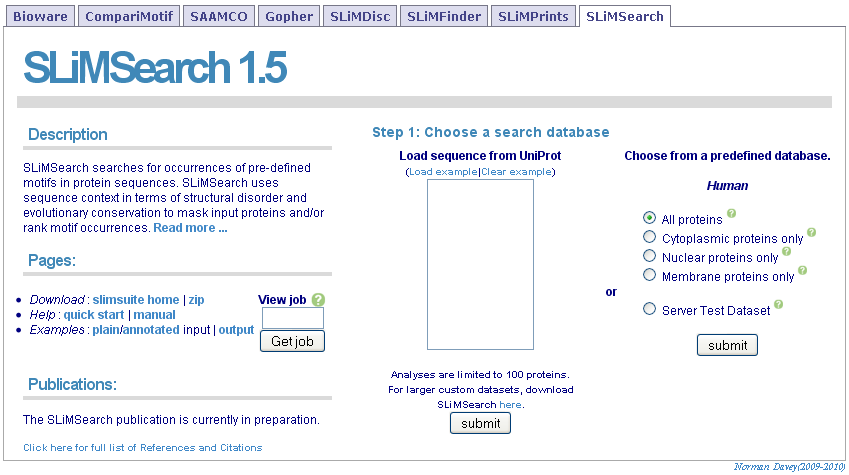
For full functionality, including SLiMChance evolutionary weighting, input a list of UniProt IDs or accession numbers and click "Get Sequences". If you have already run SLiMSearch on a dataset, you can input the job ID and click "Get job" to jump straight to the results.
Input Motifs
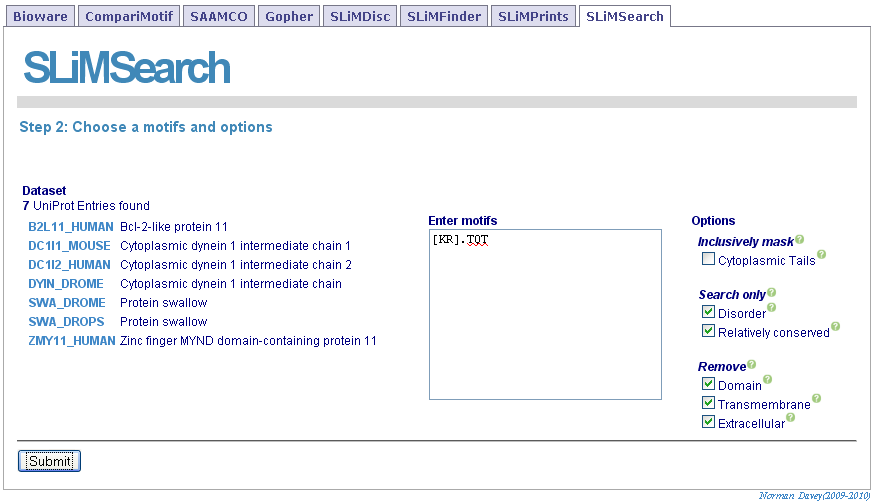
The selected sequence dataset with be shown. Motifs should be entered directly and any masking options set.
Input Dataset
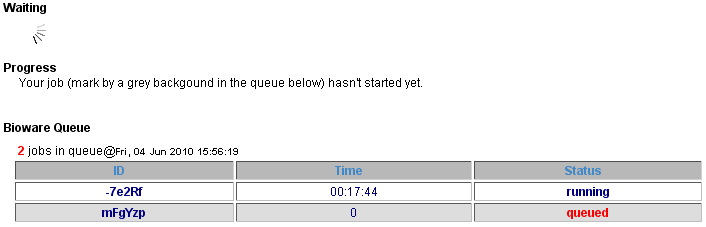
When your job is submitted, it will enter the bioware queue. The page will update once it starts running. Alternatively, the page can be bookmarked to revisit later.
Results page
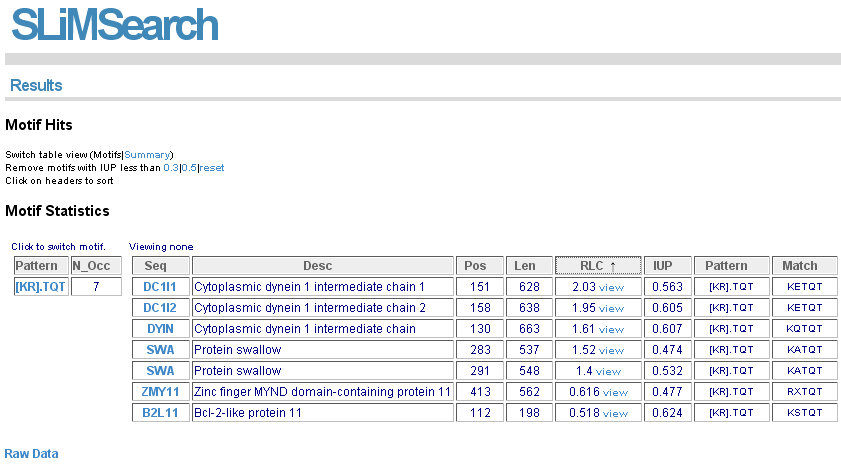
The initial results page shows motif occurrence results for returned motifs. Columns are sortable.
Protein orthologue alignments
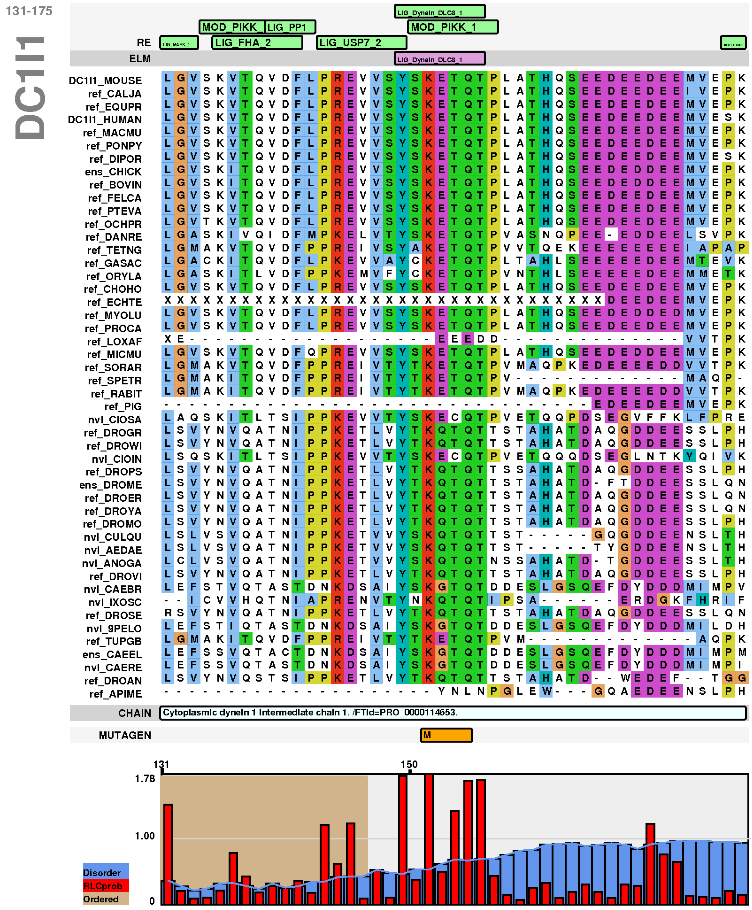
Alignments for each protein and its GOPHER orthologues around the motif of interest can be accessed by clicking on the "View" link for each occurrence. Protein disorder and RLC scores are also visualised. The region can be altered to zoom in or out as desired. These alignments can be saved by clicking on "png" or "pdf".
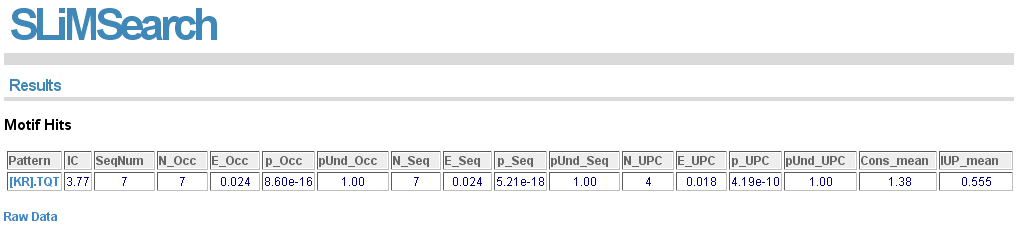
Summary results for each motif can also be visualised by clicking "Summary". All fields are sortable.