Analysing a Simple Network Example 1
For this example we will use a real protein interaction network consisting of PCNA interactors. This network was designed to give positive results to serve as an example. We are targeting the LIG_PCNA motif.
Load Data:
Data can be loaded using the standard cytoscape import function, however one attribute must contain a uniprot ID.
InteractionsAttributes
Searching for SLiM mediated Interactions
In this section we will search the loaded network for SLiMs which may be involved in binding. To do this we must first select the SLiMFinder tab and then select a suitable "Run Configuration". In this case we want to search for enrichment of sequences in interactor proteins, so we select the "Batch Interactors (Spokes)". If there is a ligand binding SLiM in multiple proteins interacting with a single protein, then this configuration may detect it.
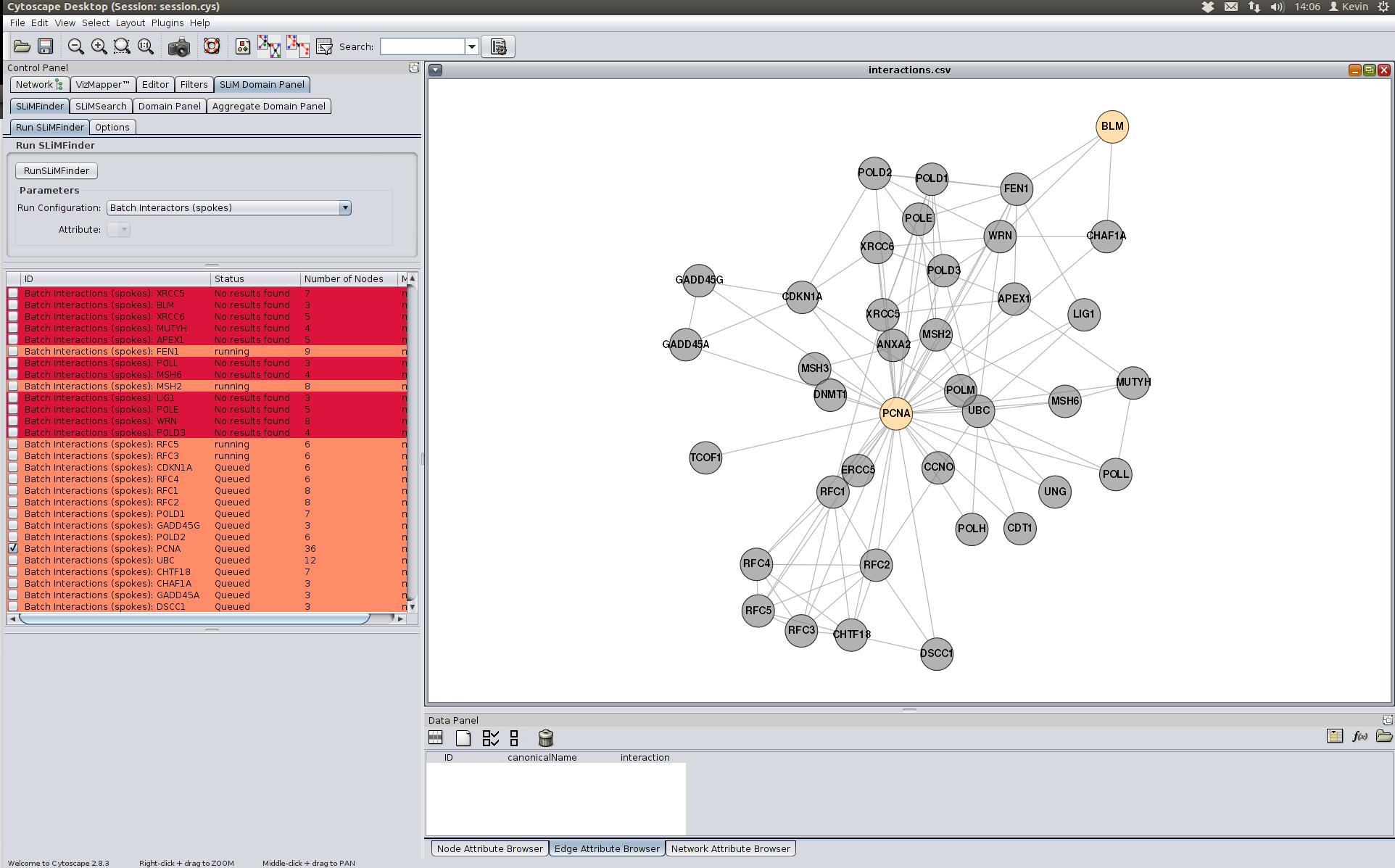
Once you have selected the correct run configuration, click the RunSLiMFinder button. This will add the run to the runs panel and they will be executed several at a time. It can take some time for this to complete as it is computationally intensive. We can see what proteins are in each protein set by clicking on the checkbox one each row of the runs table. Nodes will then be highlighted in grey as shown below.
The results for a completed run (highlighted in blue) can be shown by clicking of the row in the runs table. This will create a results table below the runs table. This can be seen in the image below where we have selected the ""Batch Interactions (spokes): PCNA" row. This shows results for interactors of PCNA.
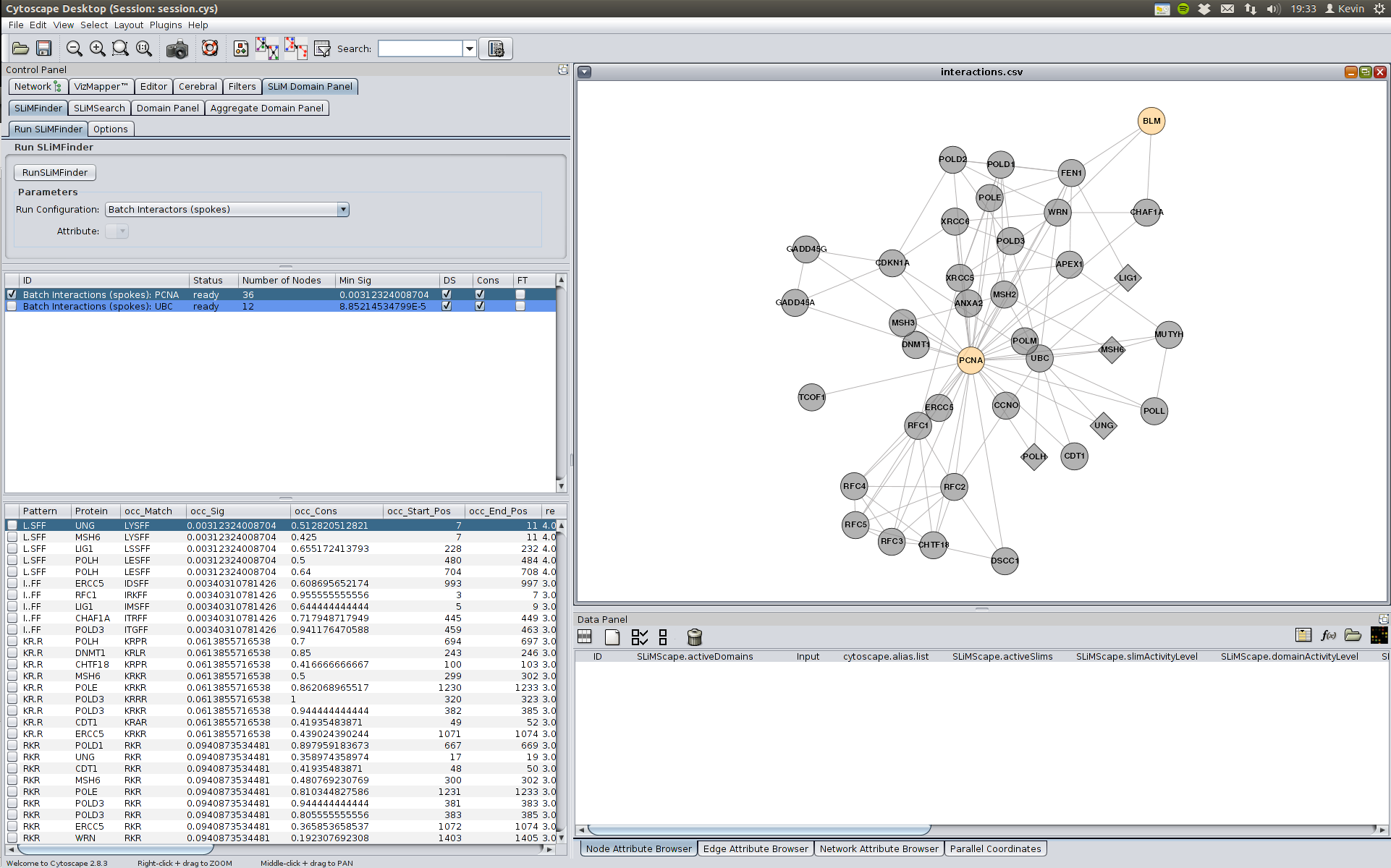
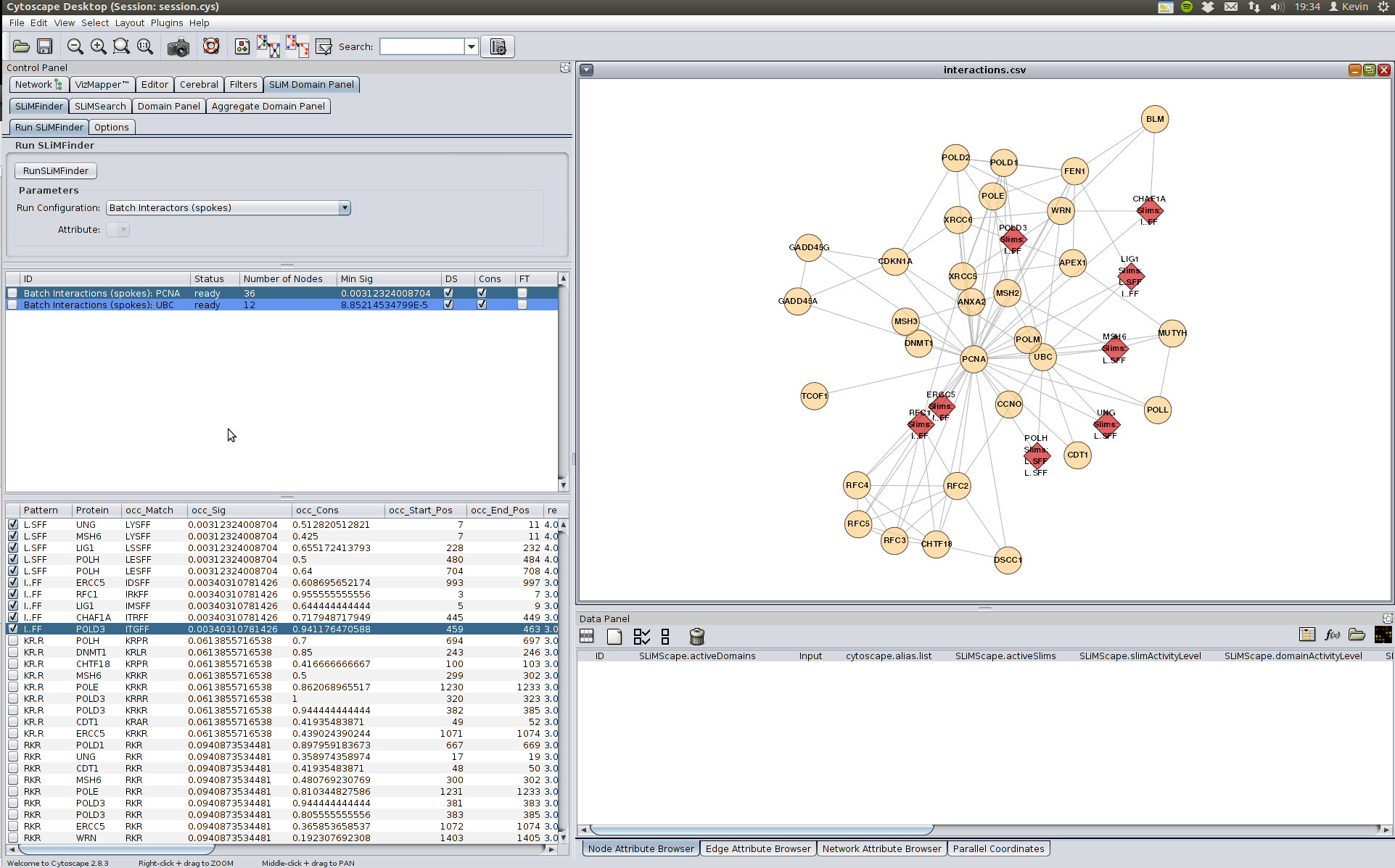
In this case there is strong enrichment (see occ_sig value) for the motif L.SFF. It is important to remember that we have performed multiple tests and therefore this could be false positive. However in this case it is a degenerate match to the LIG_PCNA motif (((^.{0,3})|(Q)).[^FHWY][ILM][^P][^FHILVWYP][HFM][FMY]..).
This motif is also present in other proteins at different levels of degeneracy. We can compare our results with the true positive LIG_PCNA instances here. It seems that many match to the known motif.
We can check if other instances of this motif exist elsewhere in the network by right clicking on one of the SLiM instances in the results panel and selecting "SLiMSearch: All Nodes". This will attempt to find instances of the potential SLiM elsewhere in the network and can add weight to existing SLiMFinder results.
We will search for the L.SFF motif.
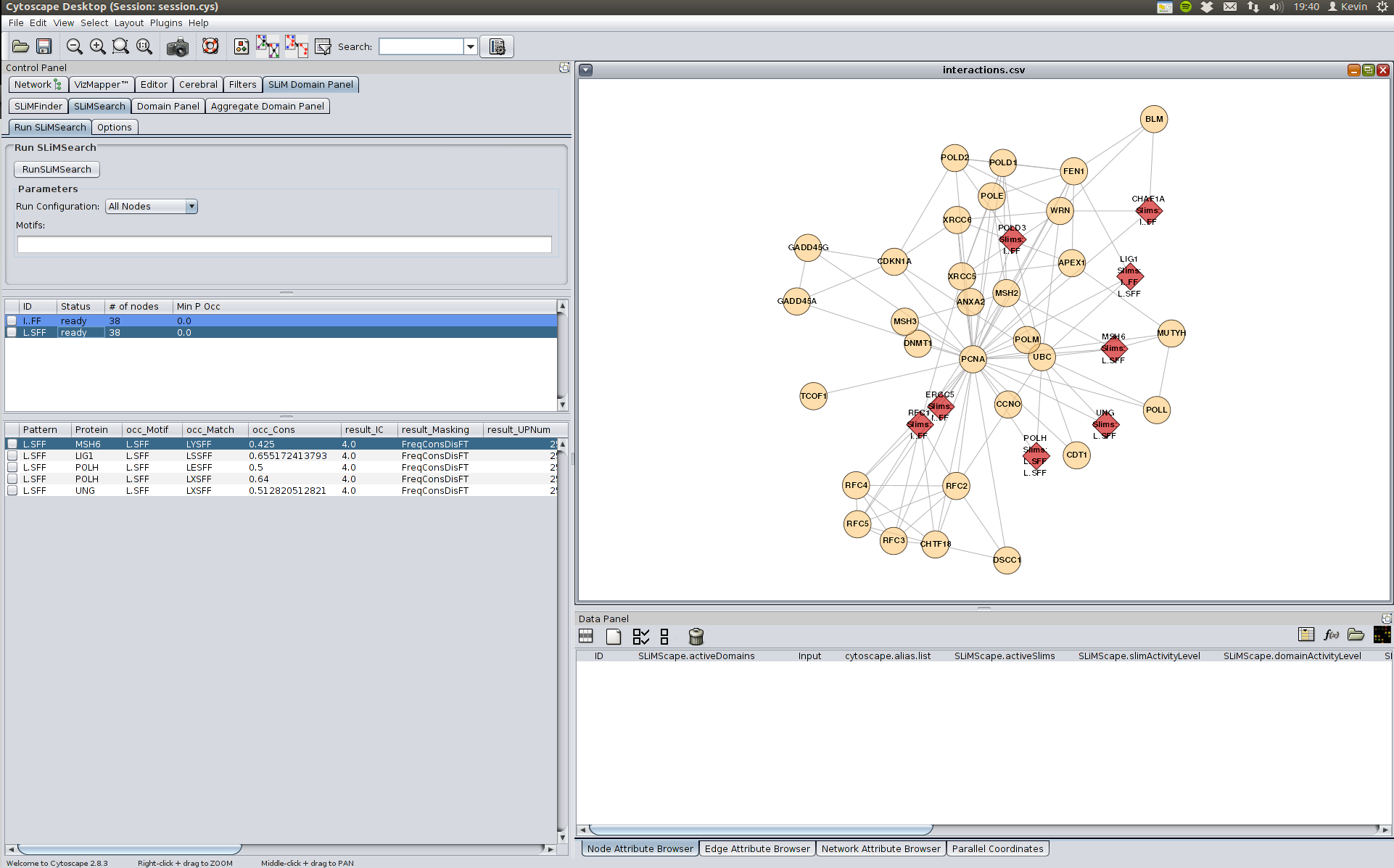
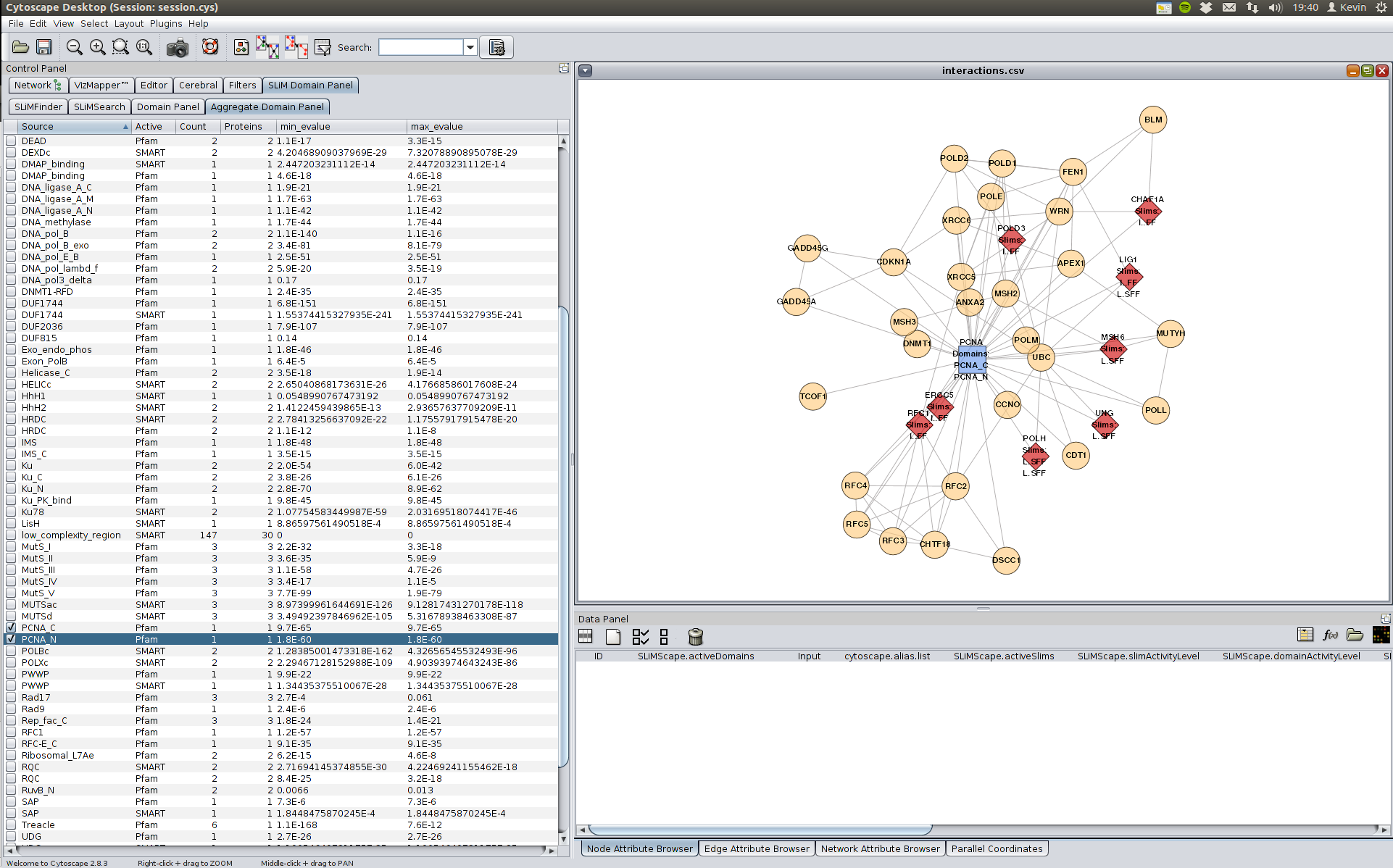
We can also look for domains which may bind this SLiM by clicking on the "Aggregate Domain Panel" tab, though in this case it is not very useful as the PCNA domains are only in PCNA and may not bind to the SLiM.